Methylated Sept9 Gene is a Sensitive Biomarker for all Stages of Colorectal Cancer
Lele Song, Yuemin Li
1Department of Radiotherapy, the Chinese PLA 309 Hospital, No.17, Heishanhu Road, HaiDian District, Beijing 100091, P.R.China Department, National Medical Center "La Raza", IMSS, Mexico City, Mexico
2BioChain (Beijing) Science and Technology, Inc, No.7A, Yongchang North Road, Economic and Technological Development Area, Beijing 100176, P.R.China
- *Corresponding Author:
- Lele Song
Department of Radiotherapy, the Chinese PLA 309 Hospital, No.17, Heishanhu Road, HaiDian District, Beijing 100091, P.R.China
Tel: 86-13240149188
E-mail: songlele@sina.com
Received date: October 11, 2015; Accepted date: October 20, 2015; Published date: October 27, 2015
Citation: Song L, Li Y. Methylated Sept9 Gene is a Sensitive Biomarker for all Stages of Colorectal Cancer. Colorec Cancer 2015, 1:1. doi: 10.21767/2471-9943.100002
Abstract
The SEPT9 gene methylation assay has been proved to be a reliable assay for CRC detection by many studies. Due to the highly sensitive properties of the assay, analysis of quantitative data is crucial for qualitative interpretation of the test results. However, different analysis methods used currently in data interpretation led to variation in test sensitivity and may affect the test effectiveness in CRC detection. Here we review the methods used in data interpretation in major clinical trials performed so far, and provide our recommendations for data interpretation in future practice.
Keywords
Septin 9; SEPT9, Septin; Methylation; Colorectal cancer; Adenoma; FOBT, FIT
Introduction
Colorectal cancer (CRC) has become the 3rd leading cause of new cancer cases in the United States in 2014[1]. The prevention of CRC should aim at early detection. However, 60%-70% of patients are found at middle or late stage CRC when they are first-time diagnosed, leading to the high mortality in CRC [2]. The low detection rate of early stage CRC is mainly due to late onset of clinical symptoms and lack of effective early detection methods. Therefore, regular screening with effective early detection methods is needed to prolong patients’ lives and reduce mortality.
Four types of test are currently available for CRC detection or screening, including fecal-based occult blood test (FOBT or FIT), tumor marker blood test, combined fecal DNA and FIT test, and imaging test (colonoscopy). The specificity or sensitivity for FOBT or FIT is not sufficient, and compliance is low due to the inconvenience in sampling and the interference of the test results by many factors [3,4]. The serum-based non-invasive tumor markers used in clinical laboratories, including carcino embryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9), are not appropriate for screening due to their low sensitivity and the lack of CRC specificity, especially for early stage CRC [5-8]. More recently, the FDA-approved fecal DNA test (Cologuard, Exact Sciences, Madison, WI, USA) exhibited high sensitivity in detecting CRC (87 %) and adenomas ≥ 1 cm (82%) [9-11]. The test includes mutation detection, methylation detection and FIT. However, the price of the test is much higher than that of the SEPT9 or FIT test, due to the combination of multiple indicators. Colonoscopy is now regarded as the gold standard for CRC diagnosis when combined with pathological examinations. However, its compliance is low due to its high costs, inconvenient preparation process, invasive procedure, and multiple complications [12]. Since these conventional CRC screening methods are either ineffective or invasive, more convenient and accurate method is needed to improve the CRC screening rate. The SEPT9 gene methylation assay, a blood-based test used specifically for CRC detection and screening, was developed and used clinically for the above purposes.
Septins are a group of scaffolding proteins that provide structural support during cell division [13]. Individual septins exist in stable six-to eight-subunit core heteromers, and the octamer contains two molecules of each of SEPT2, SEPT6, SEPT7, and SEPT9 subunits [14]. It was suggested that SEPT9 occupies a terminal position in the complex and plays a key role in subunit polymerization and the whole octamer stablization [15]. It is also critical for the final separation of daughter cells during cytokinesis [16]. Therefore, cytokinesis may be seriously affected if abnormal SEPT9 or no SEPT9 is expressed, and this could be a key factor in CRC carcinogenesis when the promoter region of the SEPT9 gene is hypermethylated and the transcription is compromised.
To date, more than ten independent clinical trials have proved the effectiveness of the assay for CRC early detection and screening. Most of these trials were case-control studies, while one screening study was performed with average-risk asymptomatic population. The sensitivity of the assay ranged from 48.2% to 95.6% in different trails, with a high specificity between 80.0% and 100.0% (Table 1) [5,17-27]. Apart from the differences in population selection, patient grouping and kit selection, the variation in sensitivity and specificity is partially due to various interpreting methods used in the trials. As the SEPT9 assay is intrinsically a quantitative assay, a dichotomized interpretation of the test results (i.e., positive or negative) was always needed to convert the quantitative data into qualitative result. This includes the setting of a cutoff value and the interpretation of data from multiple PCR reactions. This article will review the current sensitivity data from clinical trials performed so far, with specific focus on the data interpretation methods in multiple PCRs, and provide our recommendation for future clinical practice.
| Number of cases | Sensitivity | Specificity | Algorithm | Kit used |
|---|---|---|---|---|
| 309 (126 CRC, 183 control) | 72% (90/125) |
89.6% (164/183) |
2/3 | Research kit |
| 312 (133 CRC, 179 control) | 69% (92/133) |
86% (154/179) |
1/1 | Research kit |
| 245 (90 CRC, 155 control) | 73.8% (138/187) |
86.2% (282/327) |
1/3 | Research kit |
| 56.1% (105/187) |
96.6% (316/327) |
2/3 | ||
| 257 (103 CRC, 154 NED) | 67.0% (69/103) |
87.7% (135/154) |
1/2 | EpiproColon 1.0 |
| 161 (33 CRC, 34 control) | 82% (27/33) |
88% (30/34) |
1/3 | EpiproColon 1.0 |
| 73% (24/33) |
91% (31/34) |
2/3 | ||
| 144 (50 CRC, 94 control) | 90.0% (45/50) |
88.0% (83/94) |
1/3 | ARUP Lab LDT assay |
| 76.0% (38/50) |
99.1% (93/94) |
2/3 | ||
| 70.0% (35/50) |
100.0% (94/94) |
3/3 | ||
| 184 (92 CRC, 92 control) | 95.6% (88/92) |
84.8% (78/92) |
1/3 | Epi proColon 2.0 |
| 79.3% (73/92) |
99% (91/92) |
2/3 | ||
| Total 7941 (53 CRC, 1457 AA, NAA or NED) | 48.2% (standardised) |
91.5% (standardised) |
1/2 | Epi proColon 1.0 |
| 63.9% (standardised) |
88.4% (standardised) |
1/3 | ||
| Total 1544 (44 CRC, 1500 AA, small polyps or NED | 68.2% (30/44) |
80.0% (adjusted by colonoscopy) |
1/3 | Epi proColon 2.0 |
| 301 (101 CRC, 200 AA, small polyps or NED) | 73.3% (74/101) |
81.5% (163/200) |
1/3 | Epi proColon 2.0 |
| 58 (34 CRC, 24 NED) | 88.2% (30/34) |
91.7% (22/24) |
2/3 | Epi proColon 2.0 |
| 226 (135 CRC, 91 control) | 74.8% (101/135) |
96.7% (88/91) |
2/3 | Epi proColon 2.0 |
AA=Advanced Adenoma; NAA=Non-Advanced Adenoma; NED=No Evidence of Diseases.
Table 1: Sensitivity and specificity of SEPT9 gene methylation assay in CRC detection.
Detection of aberrant SEPT9 gene methylation in CRC from peripheral blood
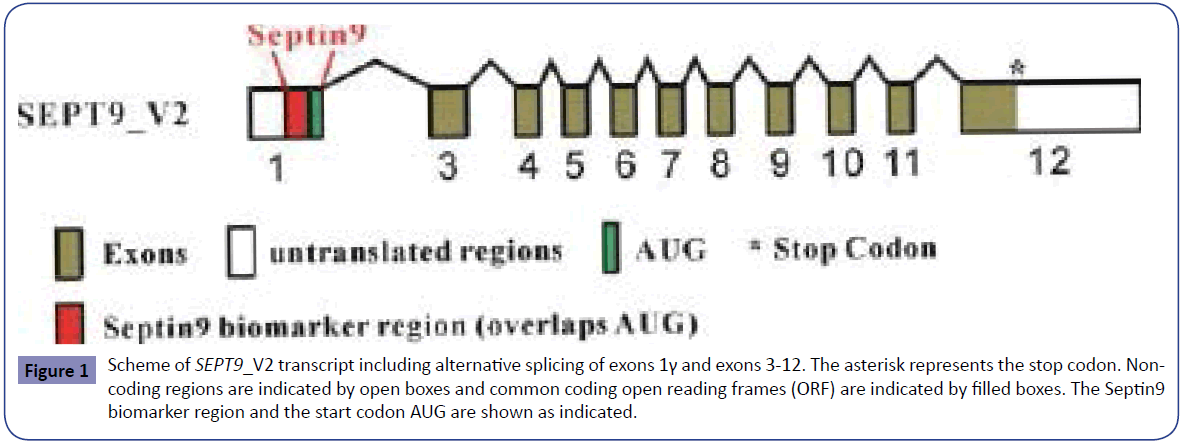
The circulating tumor DNA (ctDNA) has been suggested as a specific phenomenon and a potential source of tumor markers for tumor early detection or screening [28-30]. The SEPT9 gene methylation assay aim at detecting the aberrant methylation at the promoter region of the SEPT9 gene DNA released from CRC cells into the peripheral blood. In brief, the promoter region of the SEPT9 gene is hypermethylated and DNA of the gene is released into the peripheral circulating blood from necrotic and apoptotic cancer cells during CRC carcinogenesis, therefore, the risk of CRC can be determined by detecting the degree of DNA methylation of the specific promoter region of the SEPT9 gene in the peripheral blood [5] (Figure 1). It was shown recently that major changes in the methylation pattern of SEPT9_V2 transcript in colon adenoma and cancer tissues is confined to only one of the CpG islands, the CGI3 [18] (Figure 1). The SEPT9 gene methylation assay targets this region to specifically detect CRC (red-highlighted region in Figure 1).
Figure 1: Scheme of SEPT9_V2 transcript including alternative splicing of exons 1γ and exons 3-12. The asterisk represents the stop codon. Noncoding regions are indicated by open boxes and common coding open reading frames (ORF) are indicated by ÃÆïÃâìÃâÃÂlled boxes. The Septin9 biomarker region and the start codon AUG are shown as indicated.
The assay is composed of three steps: the extraction of plasma DNA, the bisulfite conversion and the quantitative PCR. Firstly, the cell-free circulating DNA containing trace amount of SEPT9 DNA is extracted from the peripheral blood. Samples used in this step include plasma and serum samples, whereas the extraction efficiency with plasma appears to be higher than that with serum. Secondly, bisulfite conversion is performed to allow further detection of methylation-specific changes by PCR. Unmethylated cytosine will be converted to uracil while methylated cytosine will not be converted. Finally, methylation-specific PCR is performed and specific probes are used to distinguish the methylated from unmethylated sequences. The results are presented as Ct values from PCR reactions and qualitative interpretation will be provided based on the threshold value.
Optimization of the SEPT9 assay involves multiple PCR reactions
Due to the presence of trace amount of methylated SEPT9 DNA in the peripheral blood, the detection of DNA methylation must be sensitive enough to distinguish tiny amount of methylated DNA from much higher concentration of background unmethylated genome DNA. This requires accurate design of methylation probes and optimization of the whole PCR reaction system. Since the amount of methylated SEPT9 DNA from 10 ml whole blood sample is always as low as several genome copies, multiple PCR reactions are performed to enhance the detection sensitivity. The current commercialized SEPT9 assay (e.g., Epi proColon 2.0) performs three PCR reactions and can detect as low as 2 genome copies of methylated SEPT9 per milliliter of plasma in the background of 100 ng/ml unmethylated genome DNA [24].
Three PCR reactions were chosen based on data from large amount of experiments during early development of the assay. PCR reactions more than three certainly provides better sensitivity when positive interpretation is given from at least one positive PCR reaction, however, its specificity would be low as the false positive rate increases with the increased number of reactions. The costs per assay will also increase with more consumption of reagents. Therefore, it was proved that three PCRs provide the best balance between performance and costs.
Another issue in the assay is that when three PCRs are performed, how the positive or negative interpretation can be decided. One, two or three positive PCR results are always observed when three PCR reactions are performed in parallel. The current commercialized assay (e.g., Epi proColon 2.0) interprets a positive result from at least two positive PCR reactions, and interprets a negative result from at least two negative PCR reactions. This leads to a 2 out of 3 (2/3) algorithm when interpreting the data. The reason that 2/3 algorithm was chosen is discussed in the next section.
The SEPT9 assay is sensitive for all stages of CRC regardless of algorithm
Table 1 shows the sensitivity and specificity of major clinical trials performed so far with SEPT9 gene methylation assay in CRC detection. Most of the trials were case-control or cohort trials while only one was a screening trial [23]. A big variation in sensitivity from 48.2% to 95.6% can be observed among the assays with different algorithm, while the specificity (80%-100%) remained high regardless of the variation in sensitivity.
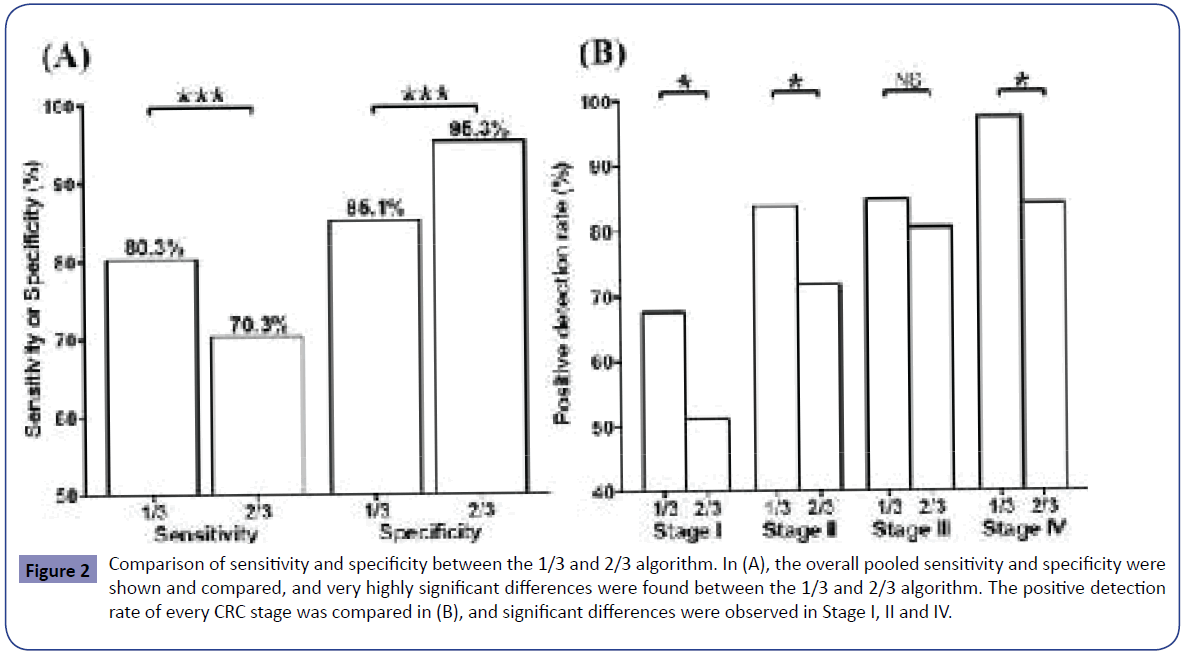
One out of three (1/3) and two out of three (2/3) were the two most common algorithms among the trials, although one out of two (1/2) and one out of one (1/1) were also observed in a couple of trials. The sensitivity data from case-control trials with the two most common algorithms is presented in (Table 2) (the screening trial was excluded from this analysis due to different trial settings [23,24]), and the overall sensitivity from each algorithm is calculated from the pooled data and compared in Figure 2(A). It can be seen that the sensitivity from the 1/3 algorithm was significantly higher (χ2=14.47, p<0.001) while the specificity from the 1/3 algorithm was significantly lower (χ2=47.35, p<0.001) than that from the 2/3 algorithm. This is a typical shift of sensitivity and specificity when algorithm is changed from 1/3 to 2/3. 1/3 algorithm led to higher sensitivity while it increased the false positive rate and therefore reduced the specificity. In contrast, 2/3 algorithm ensured high specificity at the price of sacrificing sensitivity.
| Number of cases | 1/3 algorithm | 2/3 algorithm | ||
|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |
| 309 (126 CRC, 183 control) | 72% (90/125) |
89.6% (164/183) |
||
| 245 (90 CRC, 155 control) | 73.8% (138/187) |
86.2% (282/327) |
||
| 56.1% (105/187) |
96.6% (316/327) |
|||
| 161 (33 CRC, 34 control) | 82% (27/33) |
88% (30/34) |
||
| 73% (24/33) |
91% (31/34) |
|||
| 144 (50 CRC, 94 control) | 90.0% (45/50) |
88.0% (83/94) |
||
| 76.0% (38/50) |
99.1% (93/94) |
|||
| 184 (92 CRC, 92 control) | 95.6% (88/92) |
84.8% (78/92) |
||
| 79.3% (73/92) |
99% (91/92) |
|||
| 301 (101 CRC, 200 AA, small polyps or NED) | 73.3% (74/101) |
81.5% (163/200) |
||
| 58 (34 CRC, 24 NED) | 88.2% (30/34) |
91.7% (22/24) |
||
| 226 (135 CRC, 91 control) | ÃÆãÃâââ¬Ãââ⬠| 74.8% (101/135) |
96.7% (88/91) |
|
| 80.3% (372/463) |
85.1% (636/747) |
70.3% (461/656) |
95.3% (805/845) |
|
AA=Advanced Adenoma; NAA=Non-Advanced Adenoma; NED=No Evidence of Diseases.
Table 2: Sensitivity and specificity in case-control studies when 1/3 or 2/3 algorithm was applied.
Figure 2: Comparison of sensitivity and specificity between the 1/3 and 2/3 algorithm. In (A), the overall pooled sensitivity and specificity were shown and compared, and very highly significant differences were found between the 1/3 and 2/3 algorithm. The positive detection rate of every CRC stage was compared in (B), and significant differences were observed in Stage I, II and IV.
The data of positive detection rate for each CRC stage exhibited the same trend as the overall sensitivity. Table 3 shows the individual and overall positive detection rate for each CRC stage from casecontrol studies. The overall positive detection rate of stage I (χ2=4.44,p=0.035), II (χ2=4.59,p=0.032) and IV (χ2=4.21,p=0.040) in 1/3 algorithm was significantly higher than that from the 2/3 algorithm (Table 3 and Figure 2(B)). This comparison proves that 1/3 algorithm exhibits higher sensitivity in detecting CRC in almost every stage. It can also be observed from Figure 2(B) that the SEPT9 assay was more sensitive to higher-stage CRC than lower-stage CRC, and there is a trend that the positive detection rate increased as the stage elevated.
| 1/3 algorithm | 2/3 algorithm | |||||||
| I | II | III | IV | I | II | III | IV | |
| 50.0% (11/22) |
69.4% (25/36) |
79% (42/53) |
91% (10/11) |
|||||
| 52.6% (10/19) |
75.0% (30/40) |
77.8% (21/27) |
100.0% (4/4) |
|||||
| 26.3% (5/19) |
60.0% (24/40) |
66.7% (18/27) |
75.0% (3/4) |
|||||
| 71.4% (5/7) |
90.3% (28/31) |
100.0% (7/7) |
100% (5/5) |
|||||
| 84.0% (21/25) |
100.0% (14/14) |
100.0% (35/35) |
100.0% (18/18) |
|||||
| 60.0% (15/25) |
92.8% (13/14) |
81.6% (31/35) |
77.8% (14/18) |
|||||
| 61.5% (16/26) |
80.0% (16/20) |
65.2% (15/23) |
92.3% (12/13) |
|||||
| 66.7% (12/18) |
82.6% (19/23) |
84.1% (37/44) |
100.0% (5/5) |
|||||
| Overall | 67.5% (52/77) |
83.8% (88/105) |
84.8% (78/92) |
97.5% (39/40) |
51.2% (43/84) |
71.7% (81/113) |
80.5% (128/159) |
84.2% (32/38) |
Table 3: Positive detection rate of all CRC stages when 1/3 or 2/3 algorithm was applied.
The SEPT9 assay may not be sensitive enough for adenoma detection
(Table 4) shows the positive detection rate of the SEPT9 assay in normal control and in adenoma [21-24,27]. It can be clearly seen that the positive detection rate of adenoma was much lower than that of CRC, no matter what algorithm was applied in each trial. Statistical analysis shows that significant changes in positive detection rate between normal control and adenoma can only be found in the report by Jin and colleagues [27], while no significant difference was found in other trials [21-24]. This suggests that the SEPT9 assay may not be sensitive enough as an indicator for adenoma. In Jin’s report, both NAA (χ2=6.60, p=0.010) and AA (χ2=20.03, p<0.001) exhibited significantly higher positive detection rate than the normal control. As this trial is the only one performed so far in Chinese population using Epi proColon 2.0 [27], whether or not the significant change is due to ethic difference still needs further investigation. Furthermore, a variation in the positive detection rate in normal controls among the trials can also be observed. The selection of control population could affect the positive detection rate, as we observed a trend that elder population may exhibit higher positivity rate than younger population.
| Positive detection rate | algorithm | ||
| Normal control | Adenoma | ||
| NAA | AA | ||
| 8.8% (3/34) | 7.7% (1/13) | 17.4% (4/23) | 2/3 |
| 5.7% (3/53) | 11.8% (11/93) | 9.1% (1/11) | 2/3 |
| 8.6% (80/934) | 7.7% (16/209) | 9.6% (30/314) | 1/2 |
| 21.8% (97/444) | N/A | 21.6% (134/621) | 1/3 |
| 3.3% (3/91) | 14.1% (12/85) | 27.4% (23/84) | 2/3 |
NAA=Non-Advanced Adenoma; AA=Advanced Adenoma.
Table 4: The positive detection rate of SEPT9 assay in normal control and adenoma.
It is obvious from the above discussion that 1/3 algorithm exhibited high sensitivity with low specificity, while the 2/3 algorithm exhibited lower sensitivity but higher specificity. The selection of algorithm is based on the purpose of examination using the SEPT9 assay. Currently, the SEPT9 assay is used in CRC early detection and screening. For early detection purpose in a CRC high-risk population, the exclusion of non-CRC patients may overweight the detection of all CRC patients, and specificity may therefore be more important than sensitivity in this situation, and 2/3 algorithm would be selected as the interpretation method. In contrast, for screening in average-risk population, detection of as many potential CRC patients as possible is more important than excluding non-CRC subjects, and therefore 1/3 algorithm should be applied. This recommendation is consistent with the actual applications of the commercialized SEPT9 assay. The Epi proColon 2.0 assay with 2/3 algorithm is current recommended as an auxiliary diagnosis complementary to colonoscopy in high risk population [5,19,21,22,26,27]. In contrast, 1/3 algorithm was applied in the PRESEPT study, the first screening study in averagerisk population, aiming at identifying CRC and precancerous lesions [23,24].
Conclusions
The SEPT9 assay has exhibited excellent performance in CRC early detection and screening. Selection of interpretation methods affects the sensitivity and specificity of the assay. 1/3 algorithm for CRC screening and 2/3 algorithm for early detection are recommended for the assay.
Acknowledgement
This work was supported by the capital public health project sponsored by the Beijing Municipal Science and Technology Commission. No language help, writing assistance or proof reading was used for this article.
Conflict of Interests
The first and the corresponding author of the paper, Dr. Lele Song, is currently an employee of BioChain (Beijing) Science and technology, Inc. BioChain is a collaborator of Epigenomics AG, a Germany-based company who launched the first commercial SEPT9 assay. The other author of the paper, Dr. Yuemin Li, declares no conflict of interest.
References
- Cancer Facts & Figures (2013) Atlanta, GA: American Cancer Society.
- Colorectal Cancer Facts & Figures 2011-2013. Atlanta, GA American Cancer Society.
- Imperiale TF1, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME (2004) Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population.N Engl J Med 351: 2704-2714.
- Oono Y1, Iriguchi Y, Doi Y, Tomino Y, Kishi D et al. (2010) A retrospective study of immunochemical fecal occult blood testing for colorectal cancer detection.ClinChimActa 411: 802-805.
- Tóth K1, Sipos F,Kalmár A, Patai AV, Wichmann B et al. (2012) Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers.PLoS One 7: e46000.
- Wild N1, Andres H,Rollinger W, Krause F, Dilba P, et al. (2010) A combination of serum markers for the early detection of colorectal cancer.Clin Cancer Res 16: 6111-6121.
- J.S. Chen, K.T. Chen, W.C. Fan, Yu JS, Chang YS, et al. (2010) Combined analysis of survivin autoantibody and carcinoembryonic antigen biomarkers for improved detection of colorectal cancer. ClinChem Lab Med 48: 719-725.
- Yu H1, Son GM, Joh YG (2013) The clinical significance of preoperative serum levels of carbohydrate antigen 19-9 in colorectal cancer.J Korean SurgSoc 84: 231-237.
- Ahlquist DA1, Zou H,Domanico M, Mahoney DW, Yab TC, et al. (2012) Next-generation stool DNA test accurately detects colorectal cancer and large adenomas.Gastroenterology 142: 248-256.
- Ahlquist DA1, Taylor WR, Mahoney DW, Zou H, Domanico M et al. (2012) The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia.ClinGastroenterolHepatol 10: 272-277.
- Imperiale TF1, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, et al. (2014) Multitarget stool DNA testing for colorectal-cancer screening.N Engl J Med 370: 1287-1297.
- Dominitz JA, Eisen GM, Baron TH, Goldstein JL, Hirota WK, et al. (2003) Complications of colonoscopy.GastrointestEndosc 57: 441-445.
- Longtine MS1, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, et al. (1996) The septins: roles in cytokinesis and other processes.CurrOpin Cell Biol 8: 106-119.
- Sellin ME1, Sandblad L, Stenmark S, Gullberg M (2011) Deciphering the rules governing assembly order of mammalian septin complexes.MolBiol Cell 22: 3152-3164.
- Kim MS1, Froese CD, Estey MP, Trimble WS (2011) SEPT9 occupies the terminal positions in septinoctamers and mediates polymerization-dependent functions in abscission.J Cell Biol 195: 815-826.
- Estey MP1, Di Ciano-Oliveira C, Froese CD, Bejide MT, Trimble WS (2010) Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission.J Cell Biol 191: 741-749.
- Grützmann R1, Molnar B, Pilarsky C, Habermann JK, Schlag PM, et al. (2008) Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay.PLoS One 3: e3759.
- Lofton-Day C1, Model F, Devos T, Tetzner R, Distler J, et al. (2008) DNA methylation biomarkers for blood-based colorectal cancer screening.ClinChem 54: 414-423.
- deVos T1, TetznerR, Model F, Weiss G, Schuster M, et al. (2009) Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer.ClinChem 55: 1337-1346.
- Weiss G,Rösch T (2010) Potential of a New Blood Test for Colorectal Cancer Screening –The Septin 9 Gene Biomarker. European Oncology 651–4.
- Tänzer M1, Balluff B,Distler J, Hale K, Leodolter A, et al. (2010) Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions.PLoS One 5: e9061.
- Warren JD1, Xiong W, Bunker AM, Vaughn CP, Furtado LV, et al. (2011) Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer.BMC Med 9: 133.
- Church TR1, Wandell M, Lofton-Day C, Mongin SJ, Burger M, et al. (2014) Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer.Gut 63: 317-325.
- Potter NT1, Hurban P2, White MN3, Whitlock KD3, Lofton-Day CE4, et al. (2014) Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma.ClinChem 60: 1183-1191.
- Johnson DA1, Barclay RL2, Mergener K3, Weiss G4, König T4, et al. (2014) Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study.PLoS One 9: e98238.
- Tóth K1, Wasserkort R2, Sipos F1, Kalmár A3, Wichmann B4, et al. (2014) Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA.PLoS One 9: e115415.
- Jin P1, Kang Q, Wang X, Yang L, Yu Y, et al. (2015) Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm.J GastroenterolHepatol 30: 830-833.
- Bidard FC1, Weigelt B, Reis-Filho JS (2013) Going with the flow: from circulating tumor cells to DNA.SciTransl Med 5: 207ps14.
- Pantel K1, Alix-Panabières C (2013) Real-time liquid biopsy in cancer patients: fact or fiction?Cancer Res 73: 6384-6388.
- Heitzer E1, Ulz P2, Geigl JB2 (2015) Circulating tumor DNA as a liquid biopsy for cancer.ClinChem 61: 112-123.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences